基本信息
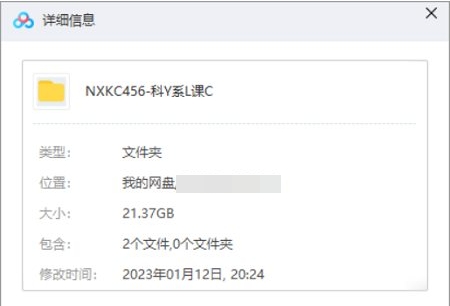
资源名称:医学方——科研系列课程
合集数目:全集
资源类型:技能学习
资源格式:MP4/AVI
资源大小:21.37GB
存储方式:百度云网盘
资料简介
解锁医学科研核心技能,助力学术突破!本套医学方——科研系列课程全集,总容量21.37GB,通过百度云网盘便捷下载,格式为MP4/AVI,专为技能学习设计。涵盖GEO/TCGA/Oncomine数据挖掘、MIMIC临床数据库、R语言作图与高级统计、WGCNA及miRNA网络构建,包含配套代码与脚本。适合医学研究者与数据分析爱好者,助力掌握数据挖掘、统计分析与基因研究,轻松提升科研能力!
资料目录
以下为资源完整目录结构(基于提供列表,规范化命名):
1. GEO、TCGA、Oncomine数据挖掘教程
1.1 数据挖掘概述.mp4
1.2 GEO在线工具应用.avi
1.3 GEO数据下载与质量分析.avi
1.4 原始数据预处理.avi
1.5 寻找差异基因及热图火山图.avi
1.6 GO富集分析.avi
1.7 KEGG分析.avi
1.8 蛋白互作网络.avi
1.9 TCGA数据下载.avi
1.10 TCGA数据整理与基因注释.avi
1.11 寻找差异基因与5年生存率.avi
1.12 Oncomine概述及Meta分析.avi
1.13 Oncomine差异分析与共表达分析.avi
1.14 非cel格式GEO芯片数据分析.mp4
1.15 数据挖掘代码
1.15.1 GEO.txt
1.15.2 TCGA.txt
1.15.3 perl/ensemblToSymbol.pl
1.15.4 perl/mRNA_merge.pl
1.15.5 perl/putFilesToOneDir.pl
1.15.6 perl/survival_time.pl
1.16 资料(脚本及源码)
1.16.1 GEO.txt
1.16.2 GEO非cel文件脚本.txt
1.16.3 TCGA.txt
1.16.4 perl/ensemblToSymbol.pl
1.16.5 perl/gb2symbol.pl
1.16.6 perl/mRNA_merge.pl
1.16.7 perl/probe2symbol.pl
1.16.8 perl/putFilesToOneDir.pl
1.16.9 perl/survival_time.pl
2. MIMIC临床数据使用入门
2.1 MIMIC临床数据库使用入门课时1-3.mp4
2.2 MIMIC临床数据库使用入门课时4-5.mp4
2.3 SQL_QUERY.docx
3. R语言可视化及绘图
3.1 par函数.mp4
3.2 散点图与盒形图.mp4
3.3 条形图与直方图.mp4
3.4 饼图与克利夫兰点图及条件图.mp4
3.5 低级绘图函数1.mp4
3.6 低级绘图函数2.mp4
3.7 图形颜色选取.mp4
3.8 ggplot2之qplot.mp4
3.9 ggplot2之美学函数.mp4
3.10 几何对象之点图.mp4
3.11 几何对象之条图.mp4
3.12 几何对象之盒形图与直方图.mp4
3.13 几何对象之线图.mp4
3.14 几何对象之标签绘制.mp4
3.15 坐标轴自定义函数.mp4
3.16 图例与标题.mp4
3.17 坐标系转换与图片分面函数.mp4
3.18 主题函数与一页多图及高质量图片导出.mp4
3.19 venn图与火山图.mp4
3.20 nomogram与heatmap.mp4
3.21 生存曲线.mp4
3.22 医学方R语言可视化配套代码.pdf
4. R语言与高级医学统计学
4.1 课时1.mp4
4.2 课时2.mp4
4.3 课时3.mp4
4.4 课时4.mp4
4.5 课时5.mp4
4.6 课时6.mp4
4.7 课时7.mp4
4.8 课时8.mp4
4.9 课时9.mp4
4.10 课时10.mp4
4.11 课时11.mp4
4.12 课时12.mp4
4.13 课时13.mp4
4.14 课时14.mp4
4.15 课时15.mp4
4.16 课时16.mp4
4.17 课时17.mp4
4.18 课时18.mp4
4.19 课时19.mp4
4.20 课时20.mp4
4.21 课时21.mp4
4.22 课时22.mp4
4.23 课时23.mp4
4.24 课时24.mp4
4.25 课时25.mp4
4.26 课时26.mp4
4.27 课时27.mp4
4.28 课时28.mp4
4.29 课时29.mp4
4.30 课时30.mp4
4.31 课时31.mp4
4.32 课时32.mp4
4.33 课时33.mp4
5. 豪斯医生数据挖掘之WGCNA详解
5.1 使用cytoscape寻找核心基因.mp4
5.2 WGCNA理论与算法1.mp4
5.3 WGCNA理论与算法2.mp4
5.4 WGCNA理论与算法3.mp4
5.5 WGCNA理论与算法4.mp4
5.6 输入文件准备.mp4
5.7 寻找基因模块.mp4
5.8 与临床信息相结合.mp4
5.9 确定核心基因.mp4
5.10 ClinicalTraits.txt
5.11 epr.txt
5.12 医学方豪斯医生WGCNA配套代码.pdf
6. 数据挖掘—miRNA调节网络的构建
6.1 TCGA概述
6.1.1 TCGA概述1.mp4
6.1.2 TCGA概述2.mp4
6.1.3 TCGA概述3.mp4
6.2 构建调控关系理论
6.2.1 调控关系理论1.mp4
6.2.2 调控关系理论2.mp4
6.2.3 调控关系理论3.mp4
6.3 WGCNA理论
6.3.1 WGCNA理论1.mp4
6.3.2 WGCNA理论2.mp4
6.3.3 WGCNA理论3.mp4
6.3.4 WGCNA理论4.mp4
6.4 数据下载
6.4.1 数据下载1.mp4
6.4.2 数据下载2.mp4
6.4.3 数据下载3.mp4
6.4.4 数据下载4.mp4
6.5 提取蛋白编码基因
6.5.1 提取蛋白编码基因1.mp4
6.5.2 提取蛋白编码基因2.mp4
6.5.3 提取蛋白编码基因3.mp4
6.5.4 提取蛋白编码基因4.mp4
6.5.5 提取蛋白编码基因5.mp4
6.6 差异分析
6.6.1 差异分析1.mp4
6.6.2 差异分析2.mp4
6.6.3 差异分析3.mp4
6.7 基于算法构建调控
6.7.1 基于算法构建调控1.mp4
6.7.2 基于算法构建调控2.mp4
6.7.3 基于算法构建调控3.mp4
6.7.4 基于算法构建调控4.mp4
6.8 cytoscape构建网络
6.8.1 cytoscape构建网络1.mp4
6.8.2 cytoscape构建网络2.mp4
6.9 数据库预测
6.9.1 数据库预测1.mp4
6.9.2 数据库预测2.mp4
6.9.3 数据库预测3.mp4
6.9.4 数据库预测4.mp4
6.10 医学方miRNA调节网络构建配套代码
6.10.1 TCGA.txt
6.10.2 DEmiRNA/diffmiRNAExp.txt
6.10.3 DEmRNA/diffmRNAExp.txt
6.10.4 merge/.Rhistory
6.10.5 merge/diffmiRNAExp.txt
6.10.6 merge/diffmRNAExp.txt
6.10.7 merge/edge.txt
6.10.8 merge/node.txt
6.10.9 merge/rmiRNA.txt
6.10.10 merge/rmRNA.txt
6.10.11 miRNA/metadata.cart.2018-01-17T14_59_36.956035.json
6.10.12 miRNA/miRNAmatrix.txt
6.10.13 miRNA/miRNA_merge.pl
6.10.14 miRNA/065fae79-0d93-4043-a332-c86f27b8bb30/793e4078-0dfd-4636-af70-c396ceae1aa4.mirbase21.mirnas.quantification.txt
6.10.15 miRNA/0a9e1555-7f9d-403f-a06f-6631173e10e6/c5c59f43-131f-4277-ad3b-479f5a0e5efe.mirbase21.mirnas.quantification.txt
6.10.16 miRNA/0d77de85-9bac-4fe6-a356-db626ddb3771/e54aa121-8eef-4bc6-bcfb-64d8f3495266.mirbase21.mirnas.quantification.txt
6.10.17 miRNA/15e40742-b59d-43b8-b904-57bf295667af/9325a28f-34ee-4ac8-8433-d7fa7a683dd8.mirbase21.mirnas.quantification.txt
6.10.18 miRNA/1658deff-1802-474c-9b88-aef6b36f101b/92190c13-4cc1-42a9-be43-1af3de505fa0.mirbase21.mirnas.quantification.txt
6.10.19 miRNA/1fca24ec-b172-4bbf-9f96-0e87be0bb252/75ea1f0a-e9c2-480f-bf40-504b9c319fd7.mirbase21.mirnas.quantification.txt
6.10.20 miRNA/2198e883-90fa-49f3-abb3-14a93c16ad97/74b2ceb4-9f5b-42fe-b711-fa41fac1d235.mirbase21.mirnas.quantification.txt
6.10.21 miRNA/39993ccb-d40e-4845-8d87-303c8b4dec44/a1545d03-4706-4494-ae4b-5757e6baff8c.mirbase21.mirnas.quantification.txt
6.10.22 miRNA/40db80b5-e815-4ae0-b128-b055924eff4d/5016bbf1-473c-4c62-92ea-35fdc043a6cd.mirbase21.mirnas.quantification.txt
6.10.23 miRNA/4f02a8fa-96eb-483d-8ae3-766756f17ce9/99b20da1-a915-4de4-ac91-a0b67f106237.mirbase21.mirnas.quantification.txt
6.10.24 miRNA/5901e47b-a554-4365-8774-3c0573062aa7/fc2f558d-4e4b-4bc7-82cd-11575fc1d55b.mirbase21.mirnas.quantification.txt
6.10.25 miRNA/5a874720-b114-495b-8ab4-32aea244b7b5/6354f55f-57dd-4030-b639-c250bf42e2d5.mirbase21.mirnas.quantification.txt
6.10.26 miRNA/5d69fc8f-cf8c-4ac0-8405-955d378a219b/6a194675-93df-474f-8f9d-c556a8153eff.mirbase21.mirnas.quantification.txt
6.10.27 miRNA/640a1b94-6187-4f04-9230-53c1bdb4cb9d/8039d5e4-8a8a-405b-b00b-049aa043654b.mirbase21.mirnas.quantification.txt
6.10.28 miRNA/74180be2-3650-48e9-ae51-e3600e897398/annotations.txt
6.10.29 miRNA/74180be2-3650-48e9-ae51-e3600e897398/f82e6b9f-c1cb-4b10-9188-5f4c5e61f711.mirbase21.mirnas.quantification.txt
6.10.30 miRNA/795ab4cc-a194-4860-bd59-7fce0e0ab9c0/1da219c5-ea21-464e-9864-2050f5ddce6a.mirbase21.mirnas.quantification.txt
6.10.31 miRNA/85997282-f179-4631-9868-01a213c354d2/8cc4f4fe-0670-4c71-b8da-3230b74d7902.mirbase21.mirnas.quantification.txt
6.10.32 miRNA/93849712-57be-4c2a-a228-503cc6de2c5b/abc5cc24-ef69-452a-8cf0-a6b92780122c.mirbase21.mirnas.quantification.txt
6.10.33 miRNA/9680279a-b495-49e3-a028-c6292b7c9b62/6d9fd070-57b0-4961-bbcd-98fb219ae8ca.mirbase21.mirnas.quantification.txt
6.10.34 miRNA/9776b29a-9b67-4a84-89a2-43dbafa15a6e/b3ac4085-b29c-4e46-b933-9d696ea1be32.mirbase21.mirnas.quantification.txt
6.10.35 miRNA/9f8123b4-6f47-4772-9cd2-598099920494/55ad3112-bc83-471f-9d65-6786ea77123d.mirbase21.mirnas.quantification.txt
6.10.36 miRNA/a5e4a61b-dbda-4399-a88f-4a28c0fa3279/e8918267-bbdc-441c-82ab-c13ce94f635e.mirbase21.mirnas.quantification.txt
6.10.37 miRNA/a687ed22-be02-4080-a937-267976ab43a7/c44e1dec-b7c4-4c2f-beda-70035466d58e.mirbase21.mirnas.quantification.txt
6.10.38 miRNA/b180b4ab-1c48-4466-ae89-eee013176a0b/fe9a5616-e3ee-49d9-abc3-239aa739104b.mirbase21.mirnas.quantification.txt
6.10.39 miRNA/b63409a1-3347-458f-8c87-981096290487/c8be7621-7e40-4d33-a77f-752af6748e90.mirbase21.mirnas.quantification.txt
6.10.40 miRNA/b9325f82-3bdc-4a54-81ea-c494132487fa/f9721fc4-b9b0-47bf-a792-e98e511ed094.mirbase21.mirnas.quantification.txt
6.10.41 miRNA/b98cf175-9133-4bec-bab5-97d87c87dcd4/d193d320-9439-41df-a184-b6a35c43aae5.mirbase21.mirnas.quantification.txt
6.10.42 miRNA/bb22a60c-ab4b-4392-9ca3-1c0f794dd06a/2c746290-217e-424d-9b1a-aac6b5176454.mirbase21.mirnas.quantification.txt
6.10.43 miRNA/c095700d-db95-412f-91ac-6ee92a4bb598/514fb76c-bfdd-48dc-ab8f-d57a67f764b9.mirbase21.mirnas.quantification.txt
6.10.44 miRNA/c2addf9e-2e7b-4c8b-84c1-b9819377fcb8/395f3a31-caa9-4ee2-b0e8-0bbe4ef51389.mirbase21.mirnas.quantification.txt
6.10.45 miRNA/c3ceb936-8fe0-4bbc-a417-6395bf544d0f/37c20413-28f6-46a4-8234-eec81b0756c2.mirbase21.mirnas.quantification.txt
6.10.46 miRNA/c536bb96-7e3b-4375-a14f-d387f778fc43/cb717142-c42a-45d3-bfff-b11f16d71806.mirbase21.mirnas.quantification.txt
6.10.47 miRNA/caab4e35-7962-4373-9922-1b393f207548/59399748-e474-4b7f-8bb2-1ebb668e3c46.mirbase21.mirnas.quantification.txt
6.10.48 miRNA/d5b74090-f9a7-4c69-92c7-4944e1d9bacd/b97c376a-241d-431d-9bb9-a336dd98caad.mirbase21.mirnas.quantification.txt
6.10.49 miRNA/d67b7afa-b10d-45b6-b47c-08e148ea9a9b/fa1cb628-2c23-44d7-a35f-3d1da147765d.mirbase21.mirnas.quantification.txt
6.10.50 miRNA/e1ac5a73-936c-44bf-bd16-48edc53434f1/680b04c9-4421-4181-bf8b-ff63ae168f71.mirbase21.mirnas.quantification.txt
6.10.51 miRNA/e7514408-e557-43a3-8b86-1a8672a24911/fa9052ca-18ff-41c1-b635-a7c8d98c2f9a.mirbase21.mirnas.quantification.txt
6.10.52 miRNA/edf54562-878d-43e8-bb62-0f875f7c4c90/ea28e0fa-2319-4d31-b2ae-5c0f58da53c4.mirbase21.mirnas.quantification.txt
6.10.53 miRNA/f0475c80-8e07-4a8a-8049-ebc35f809c52/a362f425-4bcc-4701-b89e-59c4e474df2a.mirbase21.mirnas.quantification.txt
6.10.54 miRNA/f12bd71a-e9f7-4a18-8309-7abfc0690d59/a291f6f2-62e8-498e-8c9c-6865f009eb84.mirbase21.mirnas.quantification.txt
6.10.55 miRNA/f1ecf15b-02c9-42b8-9cc2-f582ea576e6f/28ec1adc-bd57-4fc5-b63b-8a6e2213fe6e.mirbase21.mirnas.quantification.txt
6.10.56 miRNA/f25ff62c-deee-4173-ab5e-0a58730e3488/3ab052b0-080b-4b24-8674-97cdec52123f.mirbase21.mirnas.quantification.txt
6.10.57 miRNA/f32c9b2c-e2bc-4c6b-8687-3f0ea9622f08/db06db6e-9c96-4a13-a363-c8a75fcee619.mirbase21.mirnas.quantification.txt
6.10.58 miRNA/f34f743b-d4c6-479d-aee4-bd24e6ee2b92/1c2d5730-1c5b-4bc3-a85c-7f157465cc70.mirbase21.mirnas.quantification.txt
6.10.59 miRNA/ffb6ba97-1444-43c8-87f0-1af317a4a740/a91d1239-5018-447c-adf0-44b62b94a5e3.mirbase21.mirnas.quantification.txt
6.10.60 mRNA/putFilesToOneDir.pl
6.10.61 mRNA/03aee74e-4e37-4a58-a720-c90e807d2f40/annotations.txt
6.10.62 mRNA/03aee74e-4e37-4a58-a720-c90e807d2f40/be5bc6a0-9720-47ac-953e-fa8d0c32cd82.htseq.counts.gz
6.10.63 mRNA/logs/be5bc6a0-9720-47ac-953e-fa8d0c32cd82.htseq.counts.gz.parcel
6.10.64 mRNA/0d2c466e-d8b8-4b8f-9909-0be2175fa6a0/39fae157-a126-4635-a212-137065a398f9.htseq.counts.gz
6.10.65 mRNA/10d08172-48d2-49e7-b760-721163492cc1/c1071bcd-5a0c-4e09-a578-fc4b6dbe26ad.htseq.counts.gz
6.10.66 mRNA/13921de8-8580-4f26-88bb-49f71ea6e103/fbc4ffad-8a22-4531-8a3a-5761f40cfd82.htseq.counts.gz
6.10.67 mRNA/19e8fd21-f6c8-49b0-aa76-109eef46c2e9/8b20cba8-9fd5-4d56-bd02-c6f4a62767e8.htseq.counts.gz
6.10.68 mRNA/1ace0df3-9837-467e-85de-c938efda8fc8/4082f7d5-5656-476a-9aaf-36f7cea0ac55.htseq.counts.gz
6.10.69 mRNA/361cb97e-20ee-4a03-8c77-574fe10bf0d5/a8832eae-f36d-4ac5-b4c6-f8411a85d926.htseq.counts.gz
6.10.70 mRNA/3a39302b-b097-4371-a785-753c8dabb0a2/5ded64c9-6e37-413e-9e1f-8aa27b29e783.htseq.counts.gz
6.10.71 mRNA/3f34af32-53c9-4b97-8886-a3057991c466/6221d7ba-de98-418c-a341-2a6cbc3ccf23.htseq.counts.gz
6.10.72 mRNA/42994602-7f8d-4d05-a191-9bcec686aaf7/de26b3ba-4ca2-4c78-b603-4eb240315148.htseq.counts.gz
6.10.73 mRNA/42b8d463-6209-4ea0-bb01-8023a1302fa0/b6a2c03a-c8ad-41e9-8a19-8f5ac53cae9f.htseq.counts.gz
6.10.74 mRNA/494626b6-541d-417f-8ad0-1c4c44e25bef/99939497-52b7-4049-b221-12ff3332b634.htseq.counts.gz
6.10.75 mRNA/4963b9af-7d7b-42cd-b057-2f894639e59f/46fd54e8-5ab7-43f1-bb88-01a163ff121f.htseq.counts.gz
6.10.76 mRNA/4e337123-ea4c-42e5-acb9-37fce1fa7fd5/af387835-6795-467a-8103-e2083e7fb478.htseq.counts.gz
6.10.77 mRNA/4fc729ce-f4f2-492f-b305-10f51ca60972/855b771f-e894-43a8-ab38-99d0920783bd.htseq.counts.gz
6.10.78 mRNA/50e857b6-77be-4dd6-ac94-4b2186129fcb/3f3db8bd-f247-4e75-a137-7b84f5a7c6d2.htseq.counts.gz
6.10.79 mRNA/5121695c-6f2c-4fd5-b75e-25d75d1aa567/2ee0c211-17d4-4067-8520-0e1f7986793a.htseq.counts.gz
6.10.80 mRNA/533b56af-6953-4e39-b23e-ddf9a06499a5/a15f6726-8746-4176-bc5a-1f6b4a8e0ee0.htseq.counts.gz
6.10.81 mRNA/6e2031e9-df75-48df-b094-8dc6be89bf8b/c2765336-c804-4fd2-b45a-e75af2a91954.htseq.counts.gz
6.10.82 mRNA/7845948f-701e-49c5-8b76-2f0e2f0d5a76/49f80167-5bbc-4f3e-aaa0-285d029b1202.htseq.counts.gz
6.10.83 mRNA/7a5b1cfb-e4d1-4989-bfc0-6eb2afb31e2e/35c9fca2-c634-480b-962f-e6fc542d2a01.htseq.counts.gz
6.10.84 mRNA/7cde9495-e573-4b38-b89c-991076cf8cf8/38c31608-d1ad-4eb2-96f9-924f6a8d3219.htseq.counts.gz
6.10.85 mRNA/7dd1c33e-35ef-406d-b516-5b3b2a486473/fae43343-f746-4c1d-afb2-bb36967cee0f.htseq.counts.gz
6.10.86 mRNA/8499d831-d9d9-4279-8fbe-84d48a562ed0/0f0e9856-1a21-4129-a791-deb93f75fa56.htseq.counts.gz
6.10.87 mRNA/85bc7f81-51fb-4446-b12d-8741eef4acee/725eaa94-5221-4c22-bced-0c36c10c2c3b.htseq.counts.gz
6.10.88 mRNA/8e06ea1c-dbca-4b01-8f00-e4259507a77a/04df2e2a-04c6-4a8d-9bf3-b7a7a075101e.htseq.counts.gz
6.10.89 mRNA/8e31c3bb-40c8-48e2-9de1-1ee1ee31aac8/d3b8d3bc-ac3b-434f-b40b-dcd8235fb87a.htseq.counts.gz
6.10.90 mRNA/9796011a-fbab-409a-a850-bd890098c576/8a913a99-5b8d-4ebf-9d66-b93f5c92431e.htseq.counts.gz
6.10.91 mRNA/afecdda2-735c-4304-a087-ef917ad9cd5a/fc06a930-e3a1-4775-a126-610a531c655b.htseq.counts.gz
6.10.92 mRNA/b223180f-7a91-40dd-a3f1-ab423a820255/51724196-560a-47d6-adb4-5fb1ff622f21.htseq.counts.gz
6.10.93 mRNA/b48c636d-ac52-40cd-8279-0215259f8406/faac986b-ed3c-4547-a4e0-387f2d4d63b0.htseq.counts.gz
6.10.94 mRNA/b554b9e8-01c4-46ad-8909-5ada19fe5860/468c5071-fbaf-4b2e-87f4-9c75ca0096be.htseq.counts.gz
6.10.95 mRNA/b74b20b7-2797-4266-8f56-166978275576/8f00d52e-9dde-4594-87a1-8946bc7fb438.htseq.counts.gz
6.10.96 mRNA/b886634e-e0a7-459a-a2c9-20fabb779dc6/a4db2c6c-403b-4ea1-9b54-d2aae853389c.htseq.counts.gz
6.10.97 mRNA/bd31db54-5544-41e6-abab-86f2e997cfe5/6496c79a-4364-4adc-8c64-5c2fa17c0410.htseq.counts.gz
6.10.98 mRNA/be2d52c8-eb14-4b45-a65b-c1bdd6f5c4c2/4696ce44-29bf-41ea-b866-ddb17c376e94.htseq.counts.gz
6.10.99 mRNA/c32379fa-928f-4eec-b849-a5b715d1f9cb/c696175b-9427-484b-8673-49a1c321f2fa.htseq.counts.gz
6.10.100 mRNA/d9249d25-d852-4008-a71e-dd9be6cb68e9/fbdb830c-2f70-45f5-a2d6-2d8317301af3.htseq.counts.gz
6.10.101 mRNA/e4e4df95-84e9-4811-9ff9-d3bf4f3be116/908d56cc-fda7-431b-963c-e8ca753f5561.htseq.counts.gz
6.10.102 mRNA/e7a3c801-4e2f-45fd-948b-cef353403afb/85bebda7-644b-4f9b-a928-b758f4fd9495.htseq.counts.gz
6.10.103 mRNA/efb2d3a5-b73d-4895-865c-4c27d4c142bc/662bbe73-fded-44c1-abf4-bbae3f65ae3b.htseq.counts.gz
6.10.104 mRNA/f18234f6-9933-4aa7-8c17-f41f64569cf6/d5fb0f37-d8e0-4ec8-a3c5-dbc68e7b228f.htseq.counts.gz
6.10.105 mRNA/f38840ce-3838-40df-b5f2-ecd58bfe7c57/b408c617-5d6e-4328-a973-345f41db6370.htseq.counts.gz
6.10.106 mRNA/f5cf4d3b-50f0-4c39-a8c3-450436f5844a/e57fc205-c603-4fda-9524-85257817f713.htseq.counts.gz
6.10.107 mRNA/fd0ea67b-5b75-471f-be3c-a92142b91cf3/36efaf00-cc8e-437b-893d-6dcec1328022.htseq.counts.gz
6.10.108 mRNA/count/04df2e2a-04c6-4a8d-9bf3-b7a7a075101e.htseq.counts
6.10.109 mRNA/count/0f0e9856-1a21-4129-a791-deb93f75fa56.htseq.counts
6.10.110 mRNA/count/2ee0c211-17d4-4067-8520-0e1f7986793a.htseq.counts
6.10.111 mRNA/count/35c9fca2-c634-480b-962f-e6fc542d2a01.htseq.counts
6.10.112 mRNA/count/36efaf00-cc8e-437b-893d-6dcec1328022.htseq.counts
6.10.113 mRNA/count/38c31608-d1ad-4eb2-96f9-924f6a8d3219.htseq.counts
6.10.114 mRNA/count/39fae157-a126-4635-a212-137065a398f9.htseq.counts
6.10.115 mRNA/count/3f3db8bd-f247-4e75-a137-7b84f5a7c6d2.htseq.counts
6.10.116 mRNA/count/4082f7d5-5656-476a-9aaf-36f7cea0ac55.htseq.counts
6.10.117 mRNA/count/468c5071-fbaf-4b2e-87f4-9c75ca0096be.htseq.counts
6.10.118 mRNA/count/4696ce44-29bf-41ea-b866-ddb17c376e94.htseq.counts
6.10.119 mRNA/count/46fd54e8-5ab7-43f1-bb88-01a163ff121f.htseq.counts
6.10.120 mRNA/count/49f80167-5bbc-4f3e-aaa0-285d029b1202.htseq.counts
6.10.121 mRNA/count/51724196-560a-47d6-adb4-5fb1ff622f21.htseq.counts
6.10.122 mRNA/count/5ded64c9-6e37-413e-9e1f-8aa27b29e783.htseq.counts
6.10.123 mRNA/count/6221d7ba-de98-418c-a341-2a6cbc3ccf23.htseq.counts
6.10.124 mRNA/count/6496c79a-4364-4adc-8c64-5c2fa17c0410.htseq.counts
6.10.125 mRNA/count/662bbe73-fded-44c1-abf4-bbae3f65ae3b.htseq.counts
6.10.126 mRNA/count/725eaa94-5221-4c22-bced-0c36c10c2c3b.htseq.counts
6.10.127 mRNA/count/855b771f-e894-43a8-ab38-99d0920783bd.htseq.counts
6.10.128 mRNA/count/85bebda7-644b-4f9b-a928-b758f4fd9495.htseq.counts
6.10.129 mRNA/count/8a913a99-5b8d-4ebf-9d66-b93f5c92431e.htseq.counts
6.10.130 mRNA/count/8b20cba8-9fd5-4d56-bd02-c6f4a62767e8.htseq.counts
6.10.131 mRNA/count/8f00d52e-9dde-4594-87a1-8946bc7fb438.htseq.counts
6.10.132 mRNA/count/908d56cc-fda7-431b-963c-e8ca753f5561.htseq.counts
6.10.133 mRNA/count/99939497-52b7-4049-b221-12ff3332b634.htseq.counts
6.10.134 mRNA/count/a15f6726-8746-4176-bc5a-1f6b4a8e0ee0.htseq.counts
6.10.135 mRNA/count/a4db2c6c-403b-4ea1-9b54-d2aae853389c.htseq.counts
6.10.136 mRNA/count/a8832eae-f36d-4ac5-b4c6-f8411a85d926.htseq.counts
6.10.137 mRNA/count/af387835-6795-467a-8103-e2083e7fb478.htseq.counts
6.10.138 mRNA/count/b408c617-5d6e-4328-a973-345f41db6370.htseq.counts
6.10.139 mRNA/count/b6a2c03a-c8ad-41e9-8a19-8f5ac53cae9f.htseq.counts
6.10.140 mRNA/count/be5bc6a0-9720-47ac-953e-fa8d0c32cd82.htseq.counts
6.10.141 mRNA/count/c1071bcd-5a0c-4e09-a578-fc4b6dbe26ad.htseq.counts
6.10.142 mRNA/count/c2765336-c804-4fd2-b45a-e75af2a91954.htseq.counts
6.10.143 mRNA/count/c696175b-9427-484b-8673-49a1c321f2fa.htseq.counts
6.10.144 mRNA/count/d3b8d3bc-ac3b-434f-b40b-dcd8235fb87a.htseq.counts
6.10.145 mRNA/count/d5fb0f37-d8e0-4ec8-a3c5-dbc68e7b228f.htseq.counts
6.10.146 mRNA/count/de26b3ba-4ca2-4c78-b603-4eb240315148.htseq.counts
6.10.147 mRNA/count/e57fc205-c603-4fda-9524-85257817f713.htseq.counts
6.10.148 mRNA/count/faac986b-ed3c-4547-a4e0-387f2d4d63b0.htseq.counts
6.10.149 mRNA/count/fae43343-f746-4c1d-afb2-bb36967cee0f.htseq.counts
6.10.150 mRNA/count/fbc4ffad-8a22-4531-8a3a-5761f40cfd82.htseq.counts
6.10.151 mRNA/count/fbdb830c-2f70-45f5-a2d6-2d8317301af3.htseq.counts
6.10.152 mRNA/count/fc06a930-e3a1-4775-a126-610a531c655b.htseq.counts
6.10.153 mRNA/metadata.cart.2017-12-14T05_54_04.012157.json
6.10.154 mRNA/mRNAmatrix.txt
6.10.155 mRNA/mRNA_merge.pl
6.10.156 mRNA/pro/diffmRNAExp.txt
6.10.157 mRNA/pro/diffSig.xls
6.10.158 mRNA/pro/down.xls
6.10.159 mRNA/pro/edgerOut.xls
6.10.160 mRNA/pro/gtf_df.Rda
6.10.161 mRNA/pro/Homo_sapiens.GRCh38.89.chr.gtf
6.10.162 mRNA/pro/mRNAmatrix.txt
6.10.163 mRNA/pro/mRNA_exprSet_matrix.txt
6.10.164 mRNA/pro/normalizeExp.txt
6.10.165 mRNA/pro/up.xls
6.10.166 mRNA/pro/vol.pdf
6.10.167 mRNA/star/DEmiRNA.txt
6.10.168 mRNA/star/DEmiRNAp.txt
6.10.169 mRNA/star/DEmRNA.txt
6.10.170 mRNA/star/starBase_miRNA.txt
6.10.171 mRNA/target/.Rhistory
6.10.172 mRNA/target/DEmRNA.txt
6.10.173 mRNA/target/getMirnaTarget.pl
6.10.174 mRNA/target/miRDB.tsv
6.10.175 mRNA/target/miRTarBase.tsv
6.10.176 mRNA/target/result.xls
6.10.177 mRNA/target/target.txt
6.10.178 mRNA/target/TargetScan.tsv
7. 医学方医学数据挖掘精品课程
7.1 数据挖掘概述.mp4
7.2 GEO在线工具应用.mp4
7.3 GEO数据下载与质量分析.mp4
7.4 原始数据预处理.mp4
7.5 寻找差异基因及热图火山图.mp4
7.6 GO富集分析.mp4
7.7 KEGG分析.mp4
7.8 蛋白互作网络.mp4
7.9 TCGA数据下载.mp4
7.10 TCGA数据整理与基因注释.mp4
7.11 寻找差异基因与5年生存率.mp4
7.12 Oncomine概述及Meta分析.mp4
7.13 Oncomine差异分析与共表达分析.mp4
7.14 课程脚本代码.zip
8. 医学R语言快速入门与数据清洗
8.1 R语言与Rstudio简介.avi
8.2 R包安装与向量.avi
8.3 数值型与逻辑型向量.avi
8.4 逻辑表达式与字符串向量.avi
8.5 因子型变量.avi
8.6 列表与矩阵.avi
8.7 数组与初识数据框.avi
8.8 数据框.avi
8.9 数据框基本操作.avi
8.10 条件与循环.avi
8.11 自定义函数与数据读取.avi
8.12 数据读取与写出.avi
8.13 数据排序与长宽型数据转换.avi
8.14 变量因子化.avi
8.15 apply函数家族.avi
8.16 数据汇总函数.avi
8.17 plyr包.avi
8.18 dplyr包.avi
8.19 data.table包.avi
8.20 缺失值识别与处理1.avi
8.21 缺失值识别与处理2.avi
8.22 异常值与重复值处理.avi
8.23 字符串处理.avi
8.24 正则表达式.avi
8.25 stringr与stringi包.avi
8.26 时间与日期数据处理.avi
8.27 lubridate包.avi
8.28 时间序列简介.avi
8.29 时间序列分析.avi
8.30 描述性统计.avi
8.31 t检验.avi
8.32 数据变换.avi
8.33 方差分析.avi
8.34 卡方检验.avi
8.35 回归分析与模型诊断.avi
8.36 模型诊断与Logistic回归.avi
8.37 生存分析与COX回归.avi
8.38 R_CODES.rt
链接失效、侵权等,请用注册本站的邮箱发邮件至 382593094@qq.com